GSEA Desktop v2.0.x
GSEA v2.0.1 Release Notes (Jan 2007)#
Release 2.0.1 corrects two known problems in the GSEA software:
- Leading edge analysis no longer fails if your GSEA report is based on a preranked list or continuous phenotype. This problem occurred and was fixed in the user interface; no change was made to the algorithm.
- GSEA Run page includes a scroll bar to keep the Run button visible regardless of your screen resolution or window size.
Release 2.0.1 also corrects an error in the GSEA User Guide: * Changed the number of permutations recommended for gene_set permutation from 10000 to 1000.
GSEA v2.0.2 Release Notes (Feb 2007)#
Release 2.0.2 supports password protection of the GSEA ftp site. This entails no user-visible change to the GSEA application.
All chip annotation and MSigDB gene set files remain freely available from the Downloads page of the GSEA web site.
Note: User-created scripts that access files from the GSEA ftp site (for example, batch jobs that execute the GSEA command line) will now prompt the user to enter the password for the ftp site. To avoid the prompt, download the required files from the Downloads page of the GSEA web site and have the scripts reference the downloaded files. Alternatively, contact the GSEA team for the ftp site password.
GSEA v2.0.3 Release Notes (Feb 2009)#
Release 2.0.3 significantly increases the processing speed of the GSEA algorithm. Our thanks to Justin Guinney of Duke University who submitted this enhancement to the code. There is no change to the GSEA algorithm. This modification simply increases the speed of the algorithm.
Timing data: On Mac OS X version 10.5.4 (2.5 GHz, 4GB RAM), GSEA was launched with 1GB memory. We analyzed the P53_hgu95av2.gct dataset using the full C2 gene set collection and 1000 permutations. Under the previous release, the analysis took 2441 seconds. Under the current release, 662 seconds.
For What's New:
3/3/2009: Release 2.0.3 of GSEA is now available. The new release significantly increases the processing speed of the GSEA algorithm. To download the latest release, simply launch GSEA from the Downloads page. If you have questions, please contact us.
GSEA v2.0.4 Release Notes (Mar 2009)#
Release 2.0.4 significantly increases the processing speed of the algorithm. Our thanks to Justin Guinney of Duke University who submitted this enhancement to the code. There is no change to the GSEA algorithm. This modification simply increases the speed of the Java GSEA application.
Timing data: On Mac OS X version 10.5.4 (2.5 GHz, 4GB RAM), GSEA was launched with 1GB memory. We analyzed the P53_hgu95av2.gct dataset using the full C2 gene set collection and 1000 permutations. Under the previous release, the analysis took 2441 seconds. Under the current release, 662 seconds.
GSEA v2.0.5 Release Notes (Dec 2009)#
Release 2.0.5 runs under Java 1.6. There is no change to the GSEA algorithm. This modification simply supports Java 1.6.
Release 2.0.4, the previous release, runs under Java 1.5. Other than that, the two releases are the same.
GSEA v2.0.6 Release Notes (Feb 2010)#
Release 2.0.6 runs under Java 1.6. There is no change to the GSEA algorithm. This modification fixes FTP access issues observed recently.
GSEA v2.0.7 Release Notes (Sep 2010)#
In release 2.0.7, there is no change to the GSEA algorithm. However, in the MSigDB browser, you can now choose which version of MSigDB you want to load into GSEA. By default, the MSigDB browser points to v3.0 of MSigDB, and you can load this information by clicking the Load database button.
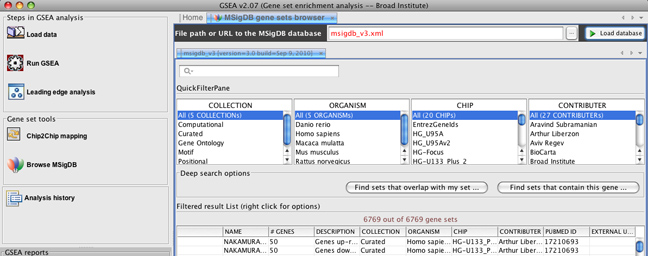
However, if you would like to load v2.5 of MSigDB, you can enter "msigdb_v2.5.xml" in the File path or URL to the MSigDB database and clicking the Load database button.
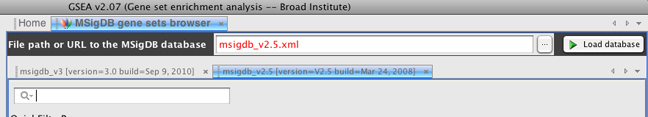
Note in the figure that both the 3.0 and 2.5 versions of MSigDB have been loaded into the browser. You can toggle between these versions by clicking their respective tabs.
GSEA v2.0.8 Release Notes (Oct 2012)#
In release 2.0.8, there is no change to the GSEA algorithm.
The changes include:
-
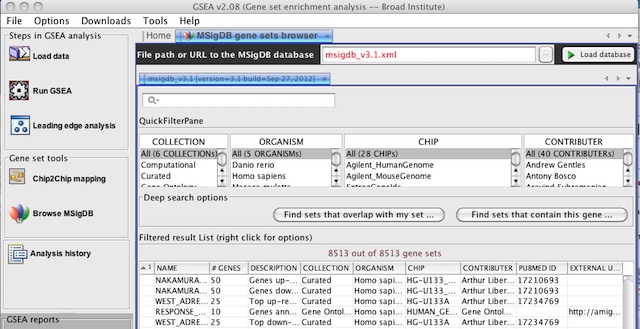
When you Browse MSigDB, by default it will now display version 3.1 of the MSigDB gene sets. To browse earlier versions of MSigDB, type msigdb_v3.xml or msigdb_v2.5.xml in the File path or URL to the MSigDB database and click the Load database button.
-
In the previous version of GSEA, there were display issues when viewing the selection menu of GMT files in the MSigDB database from the Run Gsea screen and the Chip2Chip mapping screen. These issues have been fixed. The GMT files for the latest release of MSigDB are now always displayed at the top of the list and in a bold font.
-
The latest version of MSigDB is significantly larger than previous versions and we now recommend that you run GSEA with no less than 1 GB of memory. If you run GSEA from the command line, use the -Xmx Java option to specify increased heap size. For example,
java -Xmx1024m -jar gsea2-2.08.jar.
GSEA v2.0.9 Release Notes (Dec 2012)#
GSEA v2.0.9 adds support for Java 7. With this release, the GSEA desktop software is now compatible with both Java 6 and Java 7 runtime environments.
GSEA v2.0.10 Release Notes (Jan 2013)#
GSEA v2.0.10 addresses recent issues when accessing CHIP files on our FTP server.
GSEA v2.0.12 Release Notes (Apr 2013)#
GSEA v2.0.12 includes a fix for a bug introduced in 2.0.9 for the gene list sorting mode parameter.
GSEA v2.0.13 Release Notes (Apr 2013)#
GSEA v2.0.13 has the following upgrades:
- GSEA Browser support for v4.0 MSigDB which contains new collection (C7) of the immune signatures
- GSEA determines format of input file from its extension. On some computers, system settings silently add .txt extension and do not always make it visible. This causes GSEA program to recognize these files as expression data in TXT format which in these cases might not be correct. A new feature now addresses this problem by inspecting .txt file names to see if there is a legitimate extension before .txt ending. Recognized extensions include .gct, .res, .pcl, .gmx, .gmt, and .rnk
GSEA v2.0.14 Release Notes (Jan 2014)#
GSEA v2.0.14 has the following upgrades:
- GSEA determines format of an input file from its extension. On some computers, system settings silently add .txt extension and do not always make it visible. This causes GSEA program to recognize these files as expression data in TXT format which in these cases might not be correct. In v2.0.13 we addressed this problem by inspecting .txt file names to see if there is a legitimate extension before .txt ending. Recognized extensions included .gct, .res, .pcl, .gmx, .gmt, and .rnk. In this release we have added .cls to the list of recognized file extensions.
- Improved text describing error 1001. Now reads "After pruning, none of the gene sets passed size thresholds". This communicates that tests to determine whether gene sets pass size threshold are done AFTER gene sets have been pruned to remove genes that do not appear in the gene expression data file or ranked gene list file.
- GSEA now uses anonymous FTP to access CHIP files from our FTP server.
- Updated to use the latest version (version 3.2) of apache commons math library; this eliminates reported conflict with GATK.
- Added Permissions attribute to GSEA jar file to eliminate security warning triggered by latest (7u51) Java release.